Rutin/Sulfobutylether-β-Cyclodextrin as a Promising Therapeutic Formulation for Ocular Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
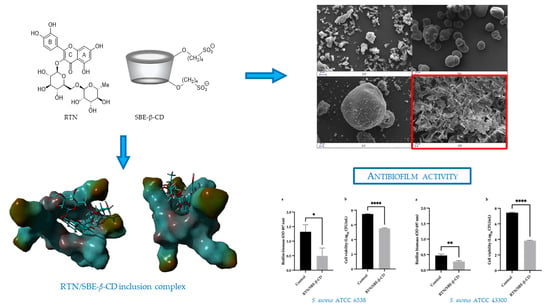
2.2. Preparation of RTN/SBE-β-CD Inclusion Complex and Physical Mixture
2.3. UV-Vis Titration
2.4. Phase-Solubility Studies
2.5. Nuclear Magnetic Resonance Experiments
2.6. Molecular Modeling Studies
2.6.1. Structure Preparation
2.6.2. Molecular Dynamics
2.6.3. Binding Free Energy Calculation
2.7. Wide-Angle X-ray Diffraction (WAXD)
2.8. Thermogravimetric Analysis (TGA)
2.9. Fourier-Transform Infrared (FT-IR) Spectroscopy
2.10. Scanning Electron Microscopy (SEM)
2.11. Determination of Water Solubility and Dissolution Profile of RTN/SBE-β-CD Inclusion Complex
2.12. In Vitro Antibacterial and Antibiofilm Activity
2.12.1. Strains
2.12.2. Susceptibility Tests
2.12.3. Effect on Biofilm Biomass and Viability
2.13. Statistical Analysis
3. Results and Discussion
3.1. Studies of RTN/SBE-β-CD Characteristics
3.1.1. UV-Vis Spectroscopy Studies
3.1.2. Phase Solubility Studies
3.1.3. NMR Investigation
3.1.4. Molecular Modeling Studies
3.1.5. WAXD, TGA, and FT-IR Analyses
3.1.6. Scanning Electron Microscopy (SEM) Analysis
3.2. Water Solubility and Dissolution Profile of RTN/SBE-β-CD Inclusion Complex
3.3. Antibacterial and Antibiofilm Activity
3.3.1. Bacterial Susceptibility
3.3.2. Antibiofilm Effect

4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ayehubizu, Z.; Mulu, W.; Biadglegne, F. Common Bacterial Causes of External Ocular Infections, Associated Risk Factors and Antibiotic Resistance among Patients at Ophthalmology Unit of Felege Hiwot Referral Hospital, Northwest Ethiopia: A Cross-Sectional Study. J. Ophthalmic Inflamm. Infect. 2021, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Ting, D.S.J.; Ho, C.S.; Deshmukh, R.; Said, D.G.; Dua, H.S. Infectious Keratitis: An Update on Epidemiology, Causative Microorganisms, Risk Factors, and Antimicrobial Resistance. Eye 2021, 35, 1084–1101. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, F.; Pignataro, D.; Di Lella, F.M.; Reibaldi, M.; Fallico, M.; Castellino, N.; Parisi, G.; Trotta, M.C.; D’Amico, M.; Santella, B.; et al. Antimicrobial Susceptibility Patterns and Resistance Trends of Staphylococcus aureus and Coagulase-Negative Staphylococci Strains Isolated from Ocular Infections. Antibiotics 2021, 10, 527. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.U.; Khurram, M.; Khattak, B.; Khan, J. Antibiotic Additive and Synergistic Action of Rutin, Morin and Quercetin against Methicillin Resistant Staphylococcus aureus. BMC Complement. Altern. Med. 2015, 15, 59. [Google Scholar] [CrossRef] [PubMed]
- Afzal, M.; Vijay, A.K.; Stapleton, F.; Willcox, M.D.P. Susceptibility of Ocular Staphylococcus aureus to Antibiotics and Multipurpose Disinfecting Solutions. Antibiotics 2021, 10, 1203. [Google Scholar] [CrossRef] [PubMed]
- Bispo, P.J.M.; Ung, L.; Chodosh, J.; Gilmore, M.S. Hospital-Associated Multidrug-Resistant MRSA Lineages Are Trophic to the Ocular Surface and Cause Severe Microbial Keratitis. Front. Public Health 2020, 8, 204. [Google Scholar] [CrossRef]
- Nithya, V.; Rathinam, S.; Siva Ganesa Karthikeyan, R.; Lalitha, P. A Ten Year Study of Prevalence, Antimicrobial Susceptibility Pattern, and Genotypic Characterization of Methicillin Resistant Staphylococcus aureus Causing Ocular Infections in a Tertiary Eye Care Hospital in South India. Infect. Genet. Evol. 2019, 69, 203–210. [Google Scholar] [CrossRef]
- Elhardt, C.; Wolf, A.; Wertheimer, C.M. Successful Treatment of Multidrug-Resistant Pseudomonas aeruginosa Keratitis with Meropenem Eye Drops—A Case Report. J. Ophthalmic Inflamm. Infect. 2023, 13, 40. [Google Scholar] [CrossRef]
- McGhee, C.N.J.; Dean, S.; Danesh-Meyer, H. Locally Administered Ocular Corticosteroids: Benefits and Risks. Drug Saf. 2002, 25, 33–55. [Google Scholar] [CrossRef]
- Kowalski, R.P.; Nayyar, S.V.; Romanowski, E.G.; Jhanji, V. Anti-Infective Treatment and Resistance Is Rarely Problematic with Eye Infections. Antibiotics 2022, 11, 204. [Google Scholar] [CrossRef]
- Shah, S.; Wozniak, R.A.F. Staphylococcus aureus and Pseudomonas aeruginosa Infectious Keratitis: Key Bacterial Mechanisms That Mediate Pathogenesis and Emerging Therapeutics. Front. Cell. Infect. Microbiol. 2023, 13, 1250257. [Google Scholar] [CrossRef] [PubMed]
- Bonincontro, G.; Scuderi, S.A.; Marino, A.; Simonetti, G. Synergistic Effect of Plant Compounds in Combination with Conventional Antimicrobials against Biofilm of Staphylococcus aureus, Pseudomonas aeruginosa, and Candida spp. Pharmaceuticals 2023, 16, 1531. [Google Scholar] [CrossRef] [PubMed]
- Di Marzio, L.; Ventura, C.A.; Cosco, D.; Paolino, D.; Di Stefano, A.; Stancanelli, R.; Tommasini, S.; Cannavà, C.; Celia, C.; Fresta, M. Nanotherapeutics for Anti-Inflammatory Delivery. J. Drug Deliv. Sci. Technol. 2016, 32, 174–191. [Google Scholar] [CrossRef]
- Memar, M.Y.; Yekani, M.; Sharifi, S.; Dizaj, S.M. Antibacterial and Biofilm Inhibitory Effects of Rutin Nanocrystals. Biointerface Res. Appl. Chem. 2022, 13, 132. [Google Scholar] [CrossRef]
- Ivanov, M.; Novović, K.; Malešević, M.; Dinić, M.; Stojković, D.; Jovčić, B.; Soković, M. Polyphenols as Inhibitors of Antibiotic Resistant Bacteria-Mechanisms Underlying Rutin Interference with Bacterial Virulence. Pharmaceuticals 2022, 15, 385. [Google Scholar] [CrossRef] [PubMed]
- Pedriali, C.A.; Fernandes, A.U.; Bernusso, L.D.C.; Polakiewicz, B. The Synthesis of a Water-Soluble Derivative of Rutin as an Antiradical Agent. Quím. Nova 2008, 31, 2147–2151. [Google Scholar] [CrossRef]
- Khalifa, T.I.; Muhtadi, F.J.; Hassan, M.M.A. Rutin. In Analytical Profiles of Drug Substances; Florey, K., Ed.; Academic Press: Cambridge, MA, USA, 1983; Volume 12, pp. 623–681. [Google Scholar]
- Gullón, B.; Lú-Chau, T.A.; Moreira, M.T.; Lema, J.M.; Eibes, G. Rutin: A Review on Extraction, Identification and Purification Methods, Biological Activities and Approaches to Enhance Its Bioavailability. Trends Food Sci. Technol. 2017, 67, 220–235. [Google Scholar] [CrossRef]
- De Gaetano, F.; Margani, F.; Barbera, V.; D’Angelo, V.; Germanò, M.P.; Pistarà, V.; Ventura, C.A. Characterization and In Vivo Antiangiogenic Activity Evaluation of Morin-Based Cyclodextrin Inclusion Complexes. Pharmaceutics 2023, 15, 2209. [Google Scholar] [CrossRef]
- De Gaetano, F.; Cristiano, M.C.; Paolino, D.; Celesti, C.; Iannazzo, D.; Pistarà, V.; Iraci, N.; Ventura, C.A. Bicalutamide Anticancer Activity Enhancement by Formulation of Soluble Inclusion Complexes with Cyclodextrins. Biomolecules 2022, 12, 1716. [Google Scholar] [CrossRef]
- De Gaetano, F.; Scala, A.; Celesti, C.; Lambertsen Larsen, K.; Genovese, F.; Bongiorno, C.; Leggio, L.; Iraci, N.; Iraci, N.; Mazzaglia, A.; et al. Amphiphilic Cyclodextrin Nanoparticles as Delivery System for Idebenone: A Preformulation Study. Molecules 2023, 28, 3023. [Google Scholar] [CrossRef]
- Musumeci, T.; Bonaccorso, A.; De Gaetano, F.; Larsen, K.L.; Pignatello, R.; Mazzaglia, A.; Puglisi, G.; Ventura, C.A. A Physico-Chemical Study on Amphiphilic Cyclodextrin/Liposomes Nanoassemblies with Drug Carrier Potential. J. Liposome Res. 2020, 30, 407–416. [Google Scholar] [CrossRef]
- Gao, S.; Zong, L.; Zhang, Y.; Zhang, Y.; Guo, X.; Guo, G.; Zhao, L.; Ye, F.; Fu, Y. Antifungal Pentachloronitrobenzene/Hydroxypropyl-Beta-Cyclodextrin Inclusion Complex Nanofibers by Electrospun with No Polymer: Fabrication and Characterization. J. Clean. Prod. 2023, 413, 137499. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, F.; Guo, G.; Xiu, Y.; Yan, H.; Zhao, L.; Gao, S.; Ye, F.; Fu, Y. Preparation and Characterization of Betulin/Methyl-Beta-Cyclodextrin Inclusion Complex Electrospun Nanofiber: Improving the Properties of Betulin. Ind. Crops Prod. 2024, 209, 117974. [Google Scholar] [CrossRef]
- Wang, Z.; Zou, W.; Liu, L.; Wang, M.; Li, F.; Shen, W. Characterization and Bacteriostatic Effects of β-Cyclodextrin/Quercetin Inclusion Compound Nanofilms Prepared by Electrospinning. Food Chem. 2021, 338, 127980. [Google Scholar] [CrossRef]
- Paladini, G.; Caridi, F.; Crupi, V.; De Gaetano, F.; Majolino, D.; Tommasini, S.; Ventura, C.A.; Venuti, V.; Stancanelli, R. Temperature-Dependent Dynamical Evolution in Coum/SBE-β-CD Inclusion Complexes Revealed by Two-Dimensional FTIR Correlation Spectroscopy (2D-COS). Molecules 2021, 26, 3749. [Google Scholar] [CrossRef]
- Matencio, A.; Hoti, G.; Monfared, Y.; Rezayat, A.; Pedrazzo, A.; Caldera, F.; Trotta, F. Cyclodextrin Monomers and Polymers for Drug Activity Enhancement. Polymers 2021, 13, 1684. [Google Scholar] [CrossRef]
- Saha, P.; Rafe, M.R. Cyclodextrin: A Prospective Nanocarrier for the Delivery of Antibacterial Agents against Bacteria That Are Resistant to Antibiotics. Heliyon 2023, 9, e19287. [Google Scholar] [CrossRef]
- Zhang, G.; Yuan, C.; Sun, Y. Effect of Selective Encapsulation of Hydroxypropyl-β-Cyclodextrin on Components and Antibacterial Properties of Star Anise Essential Oil. Molecules 2018, 23, 1126. [Google Scholar] [CrossRef]
- Haiyun, D.; Jianbin, C.; Guomei, Z.; Shaomin, S.; Jinhao, P. Preparation and Spectral Investigation on Inclusion Complex of β-Cyclodextrin with Rutin. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2003, 59, 3421–3429. [Google Scholar] [CrossRef]
- Chang, C.; Song, M.; Ma, M.; Song, J.; Cao, F.; Qin, Q. Preparation, Characterization and Molecular Dynamics Simulation of Rutin–Cyclodextrin Inclusion Complexes. Molecules 2023, 28, 955. [Google Scholar] [CrossRef]
- Calabrò, M.L.; Tommasini, S.; Donato, P.; Stancanelli, R.; Raneri, D.; Catania, S.; Costa, C.; Villari, V.; Ficarra, P.; Ficarra, R. The Rutin/β-Cyclodextrin Interactions in Fully Aqueous Solution: Spectroscopic Studies and Biological Assays. J. Pharm. Biomed. Anal. 2005, 36, 1019–1027. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Liu, B.; Zhao, J.; Thomas, D.S.; Hook, J.M. An Investigation into the Supramolecular Structure, Solubility, Stability and Antioxidant Activity of Rutin/Cyclodextrin Inclusion Complex. Food Chem. 2013, 136, 186–192. [Google Scholar] [CrossRef]
- Naeem, A.; Yu, C.; Zang, Z.; Zhu, W.; Deng, X.; Guan, Y. Synthesis and Evaluation of Rutin–Hydroxypropyl β-Cyclodextrin Inclusion Complexes Embedded in Xanthan Gum-Based (HPMC-g-AMPS) Hydrogels for Oral Controlled Drug Delivery. Antioxidants 2023, 12, 552. [Google Scholar] [CrossRef]
- Paczkowska, M.; Mizera, M.; Piotrowska, H.; Szymanowska-Powałowska, D.; Lewandowska, K.; Goscianska, J.; Pietrzak, R.; Bednarski, W.; Majka, Z.; Cielecka-Piontek, J. Complex of Rutin with β-Cyclodextrin as Potential Delivery System. PLoS ONE 2015, 10, e0120858. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, S.; Zhao, X.; Lu, Y.; Song, M.; Wu, S. Molecular Simulation and Experimental Study on the Inclusion of Rutin with β-Cyclodextrin and Its Derivative. J. Mol. Struct. 2022, 1254, 132359. [Google Scholar] [CrossRef]
- Araruna, M.K.; Brito, S.A.; Morais-Braga, M.F.; Santos, K.K.; Souza, T.M.; Leite, T.R.; Costa, J.G.; Coutinho, H.D. Coutinho Evaluation of Antibiotic & Antibiotic Modifying Activity of Pilocarpine & Rutin. Indian J. Med. Res. 2012, 135, 252–254. [Google Scholar]
- Corina, D.; Florina, B.; Iulia, P.; Cristina, D.; Rita, A.; Alexandra, P.; Virgil, P.; Hancianu, M.; Daliana, M.; Codruta, S. Rutin and Its Cyclodextrin Inclusion Complexes: Physico-Chemical Evaluation and in Vitro Activity on B164A5 Murine Melanoma Cell Line. Curr. Pharm. Biotechnol. 2018, 18, 1067–1077. [Google Scholar] [CrossRef]
- Miyake, K.; Arima, H.; Hirayama, F.; Yamamoto, M.; Horikawa, T.; Sumiyoshi, H.; Noda, S.; Uekama, K. Improvement of Solubility and Oral Bioavailability of Rutin by Complexation with 2-Hydroxypropyl-Beta-Cyclodextrin. Pharm. Dev. Technol. 2000, 5, 399–407. [Google Scholar] [CrossRef]
- Wu, M.; Song, Z.; Zhang, J. A Luminescence Study of the Interaction of Sulfobutylether-β-Cyclodextrin with Rutin. Drug Metab. Lett. 2011, 5, 259–266. [Google Scholar] [CrossRef]
- GÖZCÜ, S.; POLAT, K.H. Thermosensitive In Situ Gelling System for Dermal Drug Delivery of Rutin. Turk. J. Pharm. Sci. 2023, 20, 78–83. [Google Scholar] [CrossRef]
- Zhou, C.J.; Li, L.F.; Liu, Y.; Wen, S.P.; Guo, Y.E.; Niu, X.G. Study on the Inclusion Complex of Rutin/Sulfobutylether-β-Cyclodextrin. Adv. Mater. Res. 2012, 455–456, 1177–1181. [Google Scholar] [CrossRef]
- Das, O.; Ghate, V.M.; Lewis, S.A. Utility of Sulfobutyl Ether Beta-Cyclodextrin Inclusion Complexes in Drug Delivery: A Review. Indian J. Pharm. Sci. 2019, 81, 589–600. [Google Scholar] [CrossRef]
- Jain, A.S.; Date, A.A.; Pissurlenkar, R.R.S.; Coutinho, E.C.; Nagarsenker, M.S. Sulfobutyl Ether7 β-Cyclodextrin (SBE7 β-CD) Carbamazepine Complex: Preparation, Characterization, Molecular Modeling, and Evaluation of In Vivo Anti-Epileptic Activity. AAPS PharmSciTech 2011, 12, 1163–1175. [Google Scholar] [CrossRef]
- Sravani, A.B.; Shenoy, K.M.; Chandrika, B.; Kumar, B.H.; Kini, S.G.; Pai, K.S.R.; Lewis, S.A. Curcumin-Sulfobutyl-Ether Beta Cyclodextrin Inclusion Complex: Preparation, Spectral Characterization, Molecular Modeling, and Antimicrobial Activity. J. Biomol. Struct. Dyn. 2023, 1–16. [Google Scholar] [CrossRef]
- Higuchi, T.; Connors, K.A. Phase Solubility Techniques. Adv. Anal. Chem. Instrum. 1965, 4, 117–212. Available online: https://www.scirp.org/reference/ReferencesPapers?ReferenceID=170636 (accessed on 3 May 2023).
- Rescifina, A.; Surdo, E.; Cardile, V.; Avola, R.; Eleonora Graziano, A.C.; Stancanelli, R.; Tommasini, S.; Pistarà, V.; Ventura, C.A. Gemcitabine Anticancer Activity Enhancement by Water Soluble Celecoxib/Sulfobutyl Ether-β-Cyclodextrin Inclusion Complex. Carbohydr. Polym. 2019, 206, 792–800. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. YASARA View—Molecular Graphics for All Devices—From Smartphones to Workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef]
- Gentile, D.; Floresta, G.; Patamia, V.; Chiaramonte, R.; Mauro, G.L.; Rescifina, A.; Vecchio, M. An Integrated Pharmacophore/Docking/3D-QSAR Approach to Screening a Large Library of Products in Search of Future Botulinum Neurotoxin A Inhibitors. Int. J. Mol. Sci. 2020, 21, 9470. [Google Scholar] [CrossRef]
- Floresta, G.; Patamia, V.; Gentile, D.; Molteni, F.; Santamato, A.; Rescifina, A.; Vecchio, M. Repurposing of FDA-Approved Drugs for Treating Iatrogenic Botulism: A Paired 3D-QSAR/Docking Approach†. ChemMedChem 2020, 15, 256–262. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, Efficient Generation of High-Quality Atomic Charges. AM1-BCC Model: II. Parameterization and Validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of Multiple Amber Force Fields and Development of Improved Protein Backbone Parameters. Proteins 2006, 65, 712–725. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. New Ways to Boost Molecular Dynamics Simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert. Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Wongpituk, P.; Nutho, B.; Panman, W.; Kungwan, N.; Wolschann, P.; Rungrotmongkol, T.; Nunthaboot, N. Structural Dynamics and Binding Free Energy of Neral-Cyclodextrins Inclusion Complexes: Molecular Dynamics Simulation. Mol. Simul. 2017, 43, 1356–1363. [Google Scholar] [CrossRef]
- Azzam, A.; Shawky, R.M.; El-Mahdy, T.S. Sub-Inhibitory Concentrations of Ceftriaxone Induce Morphological Alterations and PIA-Independent Biofilm Formation in Staphylococcus aureus. Braz. J. Microbiol. 2023. [Google Scholar] [CrossRef]
- Weinstein, M.P.; Patel, J.B. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: M07-A11, 11th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; ISBN 978-1-56238-836-2. [Google Scholar]
- Cramton, S.E.; Gerke, C.; Schnell, N.F.; Nichols, W.W.; Götz, F. The Intercellular Adhesion (Ica) Locus Is Present in Staphylococcus aureus and Is Required for Biofilm Formation. Infect. Immun. 1999, 67, 5427–5433. [Google Scholar] [CrossRef]
- Mabry, T.; Markham, K.R.; Thomas, M.B. The Systematic Identification of Flavonoids; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; ISBN 978-3-642-88458-0. [Google Scholar]
- Payán-Gómez, S.A.; Flores-Holguín, N.; Pérez-Hernández, A.; Piñón-Miramontes, M.; Glossman-Mitnik, D. Computational Molecular Characterization of the Flavonoid Rutin. Chem. Cent. J. 2010, 4, 12. [Google Scholar] [CrossRef]
- Hübler, C. SupraFit—An Open Source Qt Based Fitting Application to Determine Stability Constants from Titration Experiments. Chem.–Methods 2022, 2, e202200006. [Google Scholar] [CrossRef]
- De Gaetano, F.; Marino, A.; Marchetta, A.; Bongiorno, C.; Zagami, R.; Cristiano, M.C.; Paolino, D.; Pistarà, V.; Ventura, C.A. Development of Chitosan/Cyclodextrin Nanospheres for Levofloxacin Ocular Delivery. Pharmaceutics 2021, 13, 1293. [Google Scholar] [CrossRef]
- Mura, P. Analytical Techniques for Characterization of Cyclodextrin Complexes in the Solid State: A Review. J. Pharm. Biomed. Anal. 2015, 113, 226–238. [Google Scholar] [CrossRef]
- Kringel, D.H.; Antunes, M.D.; Klein, B.; Crizel, R.L.; Wagner, R.; de Oliveira, R.P.; Dias, A.R.G.; Zavareze, E.D.R. Production, Characterization, and Stability of Orange or Eucalyptus Essential Oil/β-Cyclodextrin Inclusion Complex. J. Food Sci. 2017, 82, 2598–2605. [Google Scholar] [CrossRef]
- Miklasińska-Majdanik, M.; Kępa, M.; Wąsik, T.J.; Zapletal-Pudełko, K.; Klim, M.; Wojtyczka, R.D. The Direction of the Antibacterial Effect of Rutin Hydrate and Amikacin. Antibiotics 2023, 12, 1469. [Google Scholar] [CrossRef]
- Bernard, F.X.; Sablé, S.; Cameron, B.; Provost, J.; Desnottes, J.F.; Crouzet, J.; Blanche, F. Glycosylated Flavones as Selective Inhibitors of Topoisomerase IV. Antimicrob. Agents Chemother. 1997, 41, 992–998. [Google Scholar] [CrossRef]
- Jhanji, R.; Bhati, V.; Singh, A.; Kumar, A. Phytomolecules against Bacterial Biofilm and Efflux Pump: An in Silico and in Vitro Study. J. Biomol. Struct. Dyn. 2020, 38, 5500–5512. [Google Scholar] [CrossRef]
- Wang, Z.; Ding, Z.; Li, Z.; Ding, Y.; Jiang, F.; Liu, J. Antioxidant and Antibacterial Study of 10 Flavonoids Revealed Rutin as a Potential Antibiofilm Agent in Klebsiella pneumoniae Strains Isolated from Hospitalized Patients. Microb. Pathog. 2021, 159, 105121. [Google Scholar] [CrossRef]
- Wong, C.E.; Dolzhenko, A.V.; Lee, S.M.; Young, D.J. Cyclodextrins: A Weapon in the Fight Against Antimicrobial Resistance. J. Mol. Eng. Mater. 2017, 05, 1740006. [Google Scholar] [CrossRef]
- Diriba, K.; Kassa, T.; Alemu, Y.; Bekele, S. In Vitro Biofilm Formation and Antibiotic Susceptibility Patterns of Bacteria from Suspected External Eye Infected Patients Attending Ophthalmology Clinic, Southwest Ethiopia. Int. J. Microbiol. 2020, 2020, 8472395. [Google Scholar] [CrossRef]
- Verderosa, A.D.; Totsika, M.; Fairfull-Smith, K.E. Bacterial Biofilm Eradication Agents: A Current Review. Front. Chem. 2019, 7, 824. [Google Scholar] [CrossRef]
- Vazquez, N.M.; Mariani, F.; Torres, P.S.; Moreno, S.; Galván, E.M. Cell Death and Biomass Reduction in Biofilms of Multidrug Resistant Extended Spectrum β-Lactamase-Producing Uropathogenic Escherichia coli Isolates by 1,8-Cineole. PLoS ONE 2020, 15, e0241978. [Google Scholar] [CrossRef] [PubMed]











| Protons | RTN | RTN/SBE-β-CD | ∆δ a |
|---|---|---|---|
| 2′ | 7.67 (d) | 7.76 (d) | 0.09 |
| 5′ | 6.98 (d) | 7.05 (d) | 0.07 |
| 6′ | 7.61 (dd) | 7.71 (dd) | 0.10 |
| 6 | 6.30 (d) | 6.35 (d) | 0.05 |
| 8 | 6.49 (d) | 6.56 (d) | 1.07 |
| CH3 | 1.12 (d) | 1.20 (d) | 0.08 |
| Strains | Free RTN (μg/mL) | RTN/SBE-β-CD a (μg/mL) | LVF |
|---|---|---|---|
| S. aureus ATCC 6538 | |||
| MIC | 75 | 1.22 | 0.5 |
| MBC | 150 | 4.88 | 1 |
| S. aureus ATCC 43300 | |||
| MIC | — | 1.22 | 0.008 |
| MBC | — | 4.88 | 0.031 |
| P. aeruginosa ATCC 9027 | |||
| MIC | 150 | 39.06 | 2 |
| MBC | — | 39.06 | 62.5 |
| P. aeruginosa DSM 102273 | |||
| MIC | — | 1250 | 125 |
| MBC | — | — | 500 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Gaetano, F.; Pastorello, M.; Pistarà, V.; Rescifina, A.; Margani, F.; Barbera, V.; Ventura, C.A.; Marino, A. Rutin/Sulfobutylether-β-Cyclodextrin as a Promising Therapeutic Formulation for Ocular Infection. Pharmaceutics 2024, 16, 233. https://doi.org/10.3390/pharmaceutics16020233
De Gaetano F, Pastorello M, Pistarà V, Rescifina A, Margani F, Barbera V, Ventura CA, Marino A. Rutin/Sulfobutylether-β-Cyclodextrin as a Promising Therapeutic Formulation for Ocular Infection. Pharmaceutics. 2024; 16(2):233. https://doi.org/10.3390/pharmaceutics16020233
Chicago/Turabian StyleDe Gaetano, Federica, Martina Pastorello, Venerando Pistarà, Antonio Rescifina, Fatima Margani, Vincenzina Barbera, Cinzia Anna Ventura, and Andreana Marino. 2024. "Rutin/Sulfobutylether-β-Cyclodextrin as a Promising Therapeutic Formulation for Ocular Infection" Pharmaceutics 16, no. 2: 233. https://doi.org/10.3390/pharmaceutics16020233