Intracoronary Delivery of Porcine Cardiac Progenitor Cells Overexpressing IGF-1 and HGF in a Pig Model of Sub-Acute Myocardial Infarction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Karyotyping and Cell Proliferation Studies
2.3. Flow Cytometry
2.4. Cardiac In Vitro Differentiation of pCPC
2.5. Quantitative Real-Time PCR
2.6. RNA-Seq Analysis
2.7. pCPC Ientiviral Transduction In Vitro
2.8. Western Blot
2.9. Immunofluorescence
2.10. Co-Culture of Engineered pCPC in Decellularized Rat Cardiac Scaffolds
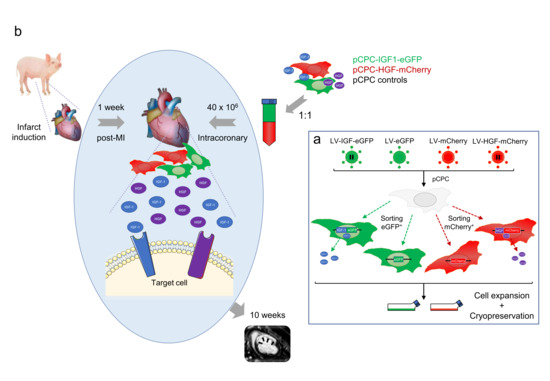
2.11. Transplantation of Engineered pCPC in Swine Myocardial Infarct Model
- -
- Group 1: Treatment group, received subpopulations A + C = pCPC-HGF-mCherry + pCPC-IGF1-eGFP (20 × 106 cells each subpopulation/animal at 2 × 106 cells/mL, total volume 20 mL).
- -
- Group 2: Control group, received subpopulations B + D = pCPC-mCherry + pCPC-eGFP (20 × 106 cells each subpopulation/animal at 2 × 106 cells/mL, total volume 20 mL).
2.12. Statistical Analysis
3. Results
3.1. Characterization of Adult pCPC
3.2. Generation and Characterization of pCPC with Forced IGF-1 or HGF Overexpression
3.3. Evaluation of the Potential Effect of IGF-1 and HGF Overexpression on pCPC Gene Expression Profile
3.4. IGF-1 Synergizes with Decellularized Rat Cardiac Scaffolds in the Promotion of Cardiogenic Differentiation of pCPC
3.5. In Vivo Evaluation of pCPC-IGF1-eGFP and pCPC-HGF-mCherry Co-Administration for the Treatment of Swine Myocardial Infarct
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ellison, G.M.; Torella, D.; Karakikes, I.; Nadal-Ginard, B. Myocyte death and renewal: Modern concepts of cardiac cellular homeostasis. Nat. Clin. Pract. Neurol. 2007, 4, S52–S59. [Google Scholar] [CrossRef]
- Ponnusamy, M.; Liu, F.; Zhang, Y.-H.; Li, R.-B.; Zhai, M.; Liu, F.; Zhou, L.-Y.; Liu, C.-Y.; Yan, K.-W.; Dong, Y.-H.; et al. Long Noncoding RNA CPR (Cardiomyocyte Proliferation Regulator) Regulates Cardiomyocyte Proliferation and Cardiac Repair. Circulation 2019, 139, 2668–2684. [Google Scholar] [CrossRef]
- Mollova, M.; Bersell, K.; Walsh, S.; Savla, J.; Das, L.T.; Park, S.-Y.; Silberstein, L.E.; dos Remedios, C.G.; Graham, D.; Colan, S.; et al. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc. Natl. Acad. Sci. USA 2013, 110, 1446–1451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabisonia, K.; Prosdocimo, G.; Aquaro, G.D.; Carlucci, L.; Zentilin, L.; Secco, I.; Ali, H.; Braga, L.; Gorgodze, N.; Bernini, F.; et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature 2019, 569, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wang, H.; Huang, X.; Li, F.; Zhu, H.; Li, Y.; He, L.; Zhang, H.; Pu, W.; Liu, K.; et al. Arterial Sca1+ Vascular Stem Cells Generate De Novo Smooth Muscle for Artery Repair and Regeneration. Cell Stem Cell 2019, 26, 81–96.e4. [Google Scholar] [CrossRef] [PubMed]
- Beltrami, A.P.; Barlucchi, L.; Torella, D.; Baker, M.; Limana, F.; Chimenti, S.; Kasahara, H.; Rota, M.; Musso, E.; Urbanek, K.; et al. Adult Cardiac Stem Cells Are Multipotent and Support Myocardial Regeneration. Cell 2003, 114, 763–776. [Google Scholar] [CrossRef] [Green Version]
- Martin-Puig, S.; Wang, Z.; Chien, K.R. Lives of a Heart Cell: Tracing the Origins of Cardiac Progenitors. Cell Stem Cell 2008, 2, 320–331. [Google Scholar] [CrossRef] [Green Version]
- Fioret, B.A.; Heimfeld, J.D.; Paik, D.T.; Hatzopoulos, A.K. Endothelial Cells Contribute to Generation of Adult Ventricular Myocytes during Cardiac Homeostasis. Cell Rep. 2014, 8, 229–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senyo, S.; Steinhauser, M.L.; Pizzimenti, C.L.; Yang, V.K.; Cai, L.; Wang, M.; Wu, T.-D.; Guerquin-Kern, J.-L.; Lechene, C.P.; Lee, R.T. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 2012, 493, 433–436. [Google Scholar] [CrossRef] [Green Version]
- Ellison, G.M.; Vicinanza, C.; Smith, A.J.; Aquila, I.; Leone, A.; Waring, C.D.; Henning, B.J.; Stirparo, G.G.; Papait, R.; Scarfò, M.; et al. Adult c-kitpos Cardiac Stem Cells Are Necessary and Sufficient for Functional Cardiac Regeneration and Repair. Cell 2013, 154, 827–842. [Google Scholar] [CrossRef] [Green Version]
- Vicinanza, C.; Aquila, I.; Scalise, M.; Cristiano, F.; Marino, F.; Cianflone, E.; Mancuso, T.; Marotta, P.; Sacco, W.; Lewis, F.; et al. Adult cardiac stem cells are multipotent and robustly myogenic: C-kit expression is necessary but not sufficient for their identification. Cell Death Differ. 2017, 24, 2101–2116. [Google Scholar] [CrossRef] [Green Version]
- Uchida, S.; de Gaspari, P.; Kostin, S.; Jenniches, K.; Kilic, A.; Izumiya, Y.; Shiojima, I.; Kreymborg, K.G.; Renz, H.; Walsh, K.; et al. Sca1-Derived Cells Are a Source of Myocardial Renewal in the Murine Adult Heart. Stem Cell Rep. 2013, 1, 397–410. [Google Scholar] [CrossRef] [Green Version]
- Malliaras, K.; Ibrahim, A.; Tseliou, E.; Liu, W.; Sun, B.; Middleton, R.C.; Seinfeld, J.; Wang, L.; Sharifi, B.G.; Marbán, E. Stimulation of endogenous cardioblasts by exogenous cell therapy after myocardial infarction. EMBO Mol. Med. 2014, 6, 760–777. [Google Scholar] [CrossRef]
- Monroe, T.O.; Hill, M.C.; Morikawa, Y.; Leach, J.P.; Heallen, T.; Cao, S.; Krijger, P.H.L.; de Laat, W.; Wehrens, X.H.T.; Rodney, G.G.; et al. YAP Partially Reprograms Chromatin Accessibility to Directly Induce Adult Cardiogenesis In Vivo. Dev. Cell 2019, 48, 765–779.e7. [Google Scholar] [CrossRef] [Green Version]
- Eschenhagen, T.; Bolli, R.; Braun, T.; Field, L.J.; Fleischmann, B.K.; Frisén, J.; Giacca, M.; Hare, J.M.; Houser, S.; Lee, R.T.; et al. Cardiomyocyte Regeneration: A consensus statement. Circulation 2017, 136, 680–686. [Google Scholar] [CrossRef]
- Malliaras, K.; Li, T.-S.; Luthringer, D.; Terrovitis, J.; Cheng, K.; Chakravarty, T.; Galang, G.; Zhang, Y.; Schoenhoff, F.; van Eyk, J.; et al. Safety and Efficacy of Allogeneic Cell Therapy in Infarcted Rats Transplanted with Mismatched Cardiosphere-Derived Cells. Circulation 2012, 125, 100–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malliaras, K.; Smith, R.R.; Kanazawa, H.; Yee, K.; Seinfeld, J.; Tseliou, E.; Dawkins, J.F.; Kreke, M.; Cheng, K.; Luthringer, D.; et al. Validation of contrast-enhanced magnetic resonance imaging to monitor regenerative efficacy after cell therapy in a porcine model of convalescent myocardial infarction. Circulation 2013, 128, 2764–2775. [Google Scholar] [CrossRef] [Green Version]
- Johnston, P.V.; Sasano, T.; Mills, K.; Evers, R.; Lee, S.-T.; Smith, R.R.; Lardo, A.C.; Lai, S.; Steenbergen, C.; Gerstenblith, G.; et al. Engraftment, Differentiation, and Functional Benefits of Autologous Cardiosphere-Derived Cells in Porcine Ischemic Cardiomyopathy. Circulation 2009, 120, 1075–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulandavelu, S.; Karantalis, V.; Fritsch, J.; Hatzistergos, K.E.; Loescher, V.Y.; McCall, F.; Wang, B.; Bagno, L.; Golpanian, S.; Wolf, A.; et al. Pim1 Kinase Overexpression Enhances ckit+ Cardiac Stem Cell Cardiac Repair Following Myocardial Infarction in Swine. J. Am. Coll. Cardiol. 2016, 68, 2454–2464. [Google Scholar] [CrossRef] [PubMed]
- Crisostomo, V.; Díaz, C.B.; Maestre, J.; Garcia-Lindo, M.; Sun, F.; Casado, J.G.; Blazquez, R.; Abad, J.L.; Palacios, I.; Rodriguez-Borlado, L.; et al. Delayed administration of allogeneic cardiac stem cell therapy for acute myocardial infarction could ameliorate adverse remodeling: Experimental study in swine. J. Transl. Med. 2015, 13, 156. [Google Scholar] [CrossRef] [Green Version]
- Bolli, R.; Tang, X.-L.; Sanganalmath, S.K.; Rimoldi, O.; Mosna, F.; Abdel-Latif, A.; Jneid, H.; Rota, M.; Leri, A.; Kajstura, J. Intracoronary Delivery of Autologous Cardiac Stem Cells Improves Cardiac Function in a Porcine Model of Chronic Ischemic Cardiomyopathy. Circulation 2013, 128, 122–131. [Google Scholar] [CrossRef] [Green Version]
- Crisostomo, V.; Baez, C.; Abad, J.L.; Sanchez, B.; Alvarez, V.; Rosado, R.; Gómez-Mauricio, G.; Gheysens, O.; Blanco-Blázquez, V.; Blazquez, R.; et al. Dose-dependent improvement of cardiac function in a swine model of acute myocardial infarction after intracoronary administration of allogeneic heart-derived cells. Stem Cell Res. Ther. 2019, 10, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makkar, R.R.; Smith, R.R.; Cheng, K.; Malliaras, K.; Thomson, L.E.J.; Berman, D.; Czer, L.S.C.; Marbán, L.; Mendizabal, A.; Johnston, P.V.; et al. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): A prospective, randomised phase 1 trial. Lancet 2012, 379, 895–904. [Google Scholar] [CrossRef] [Green Version]
- Malliaras, K.; Makkar, R.R.; Smith, R.R.; Cheng, K.; Wu, E.; Bonow, R.O.; Marbán, L.; Mendizabal, A.; Cingolani, E.; Johnston, P.V.; et al. Intracoronary Cardiosphere-Derived Cells After Myocardial Infarction. J. Am. Coll. Cardiol. 2013, 63, 110–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishigami, S.; Ohtsuki, S.; Tarui, S.; Ousaka, D.; Eitoku, T.; Kondo, M.; Okuyama, M.; Kobayashi, J.; Baba, K.; Arai, S.; et al. Intracoronary Autologous Cardiac Progenitor Cell Transfer in Patients with Hypoplastic Left Heart Syndrome: The TICAP Pro-spective Phase 1 Controlled Trial. Circ. Res. 2015, 116, 653–664. [Google Scholar] [CrossRef] [Green Version]
- Marbán, E. Breakthroughs in Cell Therapy for Heart Disease: Focus on Cardiosphere-Derived Cells. Mayo Clin. Proc. 2014, 89, 850–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chugh, A.R.; Beache, G.M.; Loughran, J.H.; Mewton, N.; Elmore, J.B.; Kajstura, J.; Pappas, P.; Tatooles, A.; Stoddard, M.F.; Lima, J.A.; et al. Administration of Cardiac Stem Cells in Patients with Ischemic Cardiomyopathy: The SCIPIO Trial. Circulation 2012, 126, S54–S64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mason, C.; Dunnill, P. Assessing the value of autologous and allogeneic cells for regenerative medicine. Regen. Med. 2009, 4, 835–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makkar, R.R.; Kereiakes, D.J.; Aguirre, F.; Kowalchuk, G.; Chakravarty, T.; Malliaras, K.; Francis, G.S.; Povsic, T.J.; Schatz, R.; Traverse, J.H.; et al. Intracoronary ALLogeneic heart STem cells to Achieve myocardial Regeneration (ALLSTAR): A randomized, placebo-controlled, double-blinded trial. Eur. Heart J. 2020, 41, 3451–3458. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Ruiz, R.; Plasencia, A.C.; Borlado, L.R.; Santos, M.E.F.; Al-Daccak, R.; Claus, P.; Palacios, I.; Sadaba, R.; Charron, D.; Bogaert, J.; et al. Rationale and Design of a Clinical Trial to Evaluate the Safety and Efficacy of Intracoronary Infusion of Allogeneic Human Cardiac Stem Cells in Patients with Acute Myocardial Infarction and Left Ventricular Dysfunction. Circ. Res. 2017, 121, 71–80. [Google Scholar] [CrossRef]
- Fernández-Avilés, F.; Sanz-Ruiz, R.; Bogaert, J.; Plasencia, A.C.; Gilaberte, I.; Belmans, A.; Santos, M.E.F.; Charron, D.; Mulet, M.; Yotti, R.; et al. Safety and Efficacy of Intracoronary Infusion of Allogeneic Human Cardiac Stem Cells in Patients With ST-Segment Elevation Myocardial Infarction and Left Ventricular Dysfunction. Circ. Res. 2018, 123, 579–589. [Google Scholar] [CrossRef]
- Matsuura, K.; Honda, A.; Nagai, T.; Fukushima, N.; Iwanaga, K.; Tokunaga, M.; Shimizu, T.; Okano, T.; Kasanuki, H.; Hagiwara, N.; et al. Transplantation of cardiac progenitor cells ameliorates cardiac dysfunction after myocardial infarction in mice. J. Clin. Investig. 2009, 119, 2204–2217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.-L.; Rokosh, G.; Sanganalmath, S.K.; Yuan, F.; Sato, H.; Mu, J.; Dai, S.; Li, C.; Chen, N.; Peng, Y.; et al. Intracoronary Administration of Cardiac Progenitor Cells Alleviates Left Ventricular Dysfunction in Rats With a 30-Day-Old Infarction. Circulation 2010, 121, 293–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, K.U.; Guo, Y.; Li, Q.-H.; Cao, P.; Al-Maqtari, T.; Vajravelu, B.N.; Du, J.; Book, M.J.; Zhu, X.; Nong, Y.; et al. c-kit+ Cardiac Stem Cells Alleviate Post-Myocardial Infarction Left Ventricular Dysfunction Despite Poor Engraftment and Negligible Retention in the Recipient Heart. PLoS ONE 2014, 9, e96725. [Google Scholar] [CrossRef] [Green Version]
- Maxeiner, H.; Krehbiehl, N.; Müller, A.; Woitasky, N.; Akintürk, H.; Müller, M.; Weigand, M.A.; Abdallah, Y.; Kasseckert, S.; Schreckenberg, R.; et al. New insights into paracrine mechanisms of human cardiac progenitor cells. Eur. J. Heart Fail. 2010, 12, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Barile, L.; Cervio, E.; Lionetti, V.; Milano, G.; Ciullo, A.; Biemmi, V.; Bolis, S.; Altomare, C.; Matteucci, M.; Di Silvestre, D.; et al. Cardioprotection by cardiac progenitor cell-secreted exosomes: Role of pregnancy-associated plasma protein-A. Cardiovasc. Res. 2018, 114, 992–1005. [Google Scholar] [CrossRef] [Green Version]
- Torán, J.L.; Aguilar, S.; Lopez, J.A.; Torroja, C.; Quintana, J.A.; Santiago, C.; Abad, J.L.; Gomes-Alves, P.; Gonzalez, A.; Bernal, J.; et al. CXCL6 is an important paracrine factor in the pro-angiogenic human cardiac progenitor-like cell secretome. Sci. Rep. 2017, 7, 12490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellison-Hughes, G.M.; Madeddu, P. Exploring pericyte and cardiac stem cell secretome unveils new tactics for drug discovery. Pharmacol. Ther. 2016, 171, 1–12. [Google Scholar] [CrossRef]
- Chimenti, I.; Smith, R.R.; Li, T.-S.; Gerstenblith, G.; Messina, E.; Giacomello, A.; Marbán, E. Relative Roles of Direct Regeneration Versus Paracrine Effects of Human Cardiosphere-Derived Cells Transplanted Into Infarcted Mice. Circ. Res. 2010, 106, 971–980. [Google Scholar] [CrossRef]
- Zeng, B.; Liu, L.; Wang, S.; Dai, Z. ILK regulates MSCs survival and angiogenesis partially through AKT and mTOR signaling pathways. Acta Histochem. 2017, 119, 400–406. [Google Scholar] [CrossRef]
- Weinberger, F.; Mannhardt, I.; Eschenhagen, T. Engineering Cardiac Muscle Tissue. Circ. Res. 2017, 120, 1487–1500. [Google Scholar] [CrossRef] [PubMed]
- Gallo, S.; Sala, V.; Gatti, S.; Crepaldi, T. HGF/Met Axis in Heart Function and Cardioprotection. Biomedicines 2014, 2, 247–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, H.; Nakamura, T.; Matsumoto, K.; Sawa, Y.; Matsuda, H.; Nakamura, T. A potential cardioprotective role of hepatocyte growth factor in myocardial infarction in rats. Cardiovasc. Res. 2001, 51, 41–50. [Google Scholar] [CrossRef]
- Kitta, K.; Day, R.M.; Ikeda, T.; Suzuki, Y.J. Hepatocyte growth factor protects cardiac myocytes against oxidative stress-induced apoptosis. Free Radic. Biol. Med. 2001, 31, 902–910. [Google Scholar] [CrossRef]
- Liu, J.; Wu, P.; Wang, Y.; Du, Y.; A, N.; Liu, S.; Zhang, Y.; Zhou, N.; Xu, Z.-H.; Yang, Z. Ad-HGF improves the cardiac remodeling of rat following myocardial infarction by upregulating autophagy and necroptosis and inhibiting apoptosis. Am. J. Transl. Res. 2016, 8, 4605–4627. [Google Scholar]
- Tao, Z.; Chen, B.; Zhao, Y.; Chen, H.; Wang, L.; Yong, Y.; Cao, K.; Yu, Q.; Ke, D.; Wang, H.; et al. HGF percutaneous endocardial injection induces cardiomyocyte proliferation and rescues cardiac function in pigs. J. Biomed. Res. 2010, 24, 198–206. [Google Scholar] [CrossRef] [Green Version]
- Sadat, S.; Gehmert, S.; Song, Y.-H.; Yen, Y.; Bai, X.; Gaiser, S.; Klein, H.; Alt, E. The cardioprotective effect of mesenchymal stem cells is mediated by IGF-I and VEGF. Biochem. Biophys. Res. Commun. 2007, 363, 674–679. [Google Scholar] [CrossRef]
- Engels, M.C.; Rajarajan, K.; Feistritzer, R.; Sharma, A.; Nielsen, U.B.; Schalij, M.J.; de Vries, A.A.; Pijnappels, D.A.; Wu, S.M. Insulin-Like Growth Factor Promotes Cardiac Lineage Induction In Vitro by Selective Expansion of Early Mesoderm. Stem Cells 2014, 32, 1493–1502. [Google Scholar] [CrossRef] [Green Version]
- Troncoso, R.; Ibarra, C.; Vicencio, J.M.; Jaimovich, E.; Lavandero, S. New insights into IGF-1 signaling in the heart. Trends Endocrinol. Metab. 2014, 25, 128–137. [Google Scholar] [CrossRef]
- Lin, M.; Liu, X.; Zheng, H.; Huang, X.; Wu, Y.; Huang, A.; Zhu, H.; Hu, Y.; Mai, W.; Huang, Y. IGF-1 enhances BMSC viability, migration, and anti-apoptosis in myocardial infarction via secreted frizzled-related protein 2 pathway. Stem Cell Res. Ther. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Bagno, L.L.; Carvalho, D.; Mesquita, F.; Louzada, R.A.; Andrade, B.; Kasai-Brunswick, T.H.; Lago, V.M.; Suhet, G.; Cipitelli, D.; Werneck-De-Castro, J.P.; et al. Sustained IGF-1 Secretion by Adipose-Derived Stem Cells Improves Infarcted Heart Function. Cell Transplant. 2016, 25, 1609–1622. [Google Scholar] [CrossRef] [Green Version]
- Báez-Díaz, C.; Blanco-Blázquez, V.; Sánchez-Margallo, F.-M.; Bayes-Genis, A.; González, I.; Abad, A.; Steendam, R.; Franssen, O.; Palacios, I.; Sánchez, B.; et al. Microencapsulated Insulin-Like Growth Factor-1 therapy improves cardiac function and reduces fibrosis in a porcine acute myocardial infarction model. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, L.-L.; Du, W.; Yu, Y.-C.; Ju, W.-Z.; Man, Y.-L.; Li, X.-R.; Chen, Y.; Wang, Z.-D.; Gu, W.-J.; et al. Hepatocyte Growth Factor Modification Enhances the Anti-Arrhythmic Properties of Human Bone Marrow-Derived Mesenchymal Stem Cells. PLoS ONE 2014, 9, e111246. [Google Scholar] [CrossRef]
- Sonnenberg, S.B.; Rane, A.A.; Liu, C.J.; Rao, N.; Agmon, G.; Suarez, S.; Wang, R.; Munoz, A.; Bajaj, V.; Zhang, S.; et al. Delivery of an engineered HGF fragment in an extracellular matrix-derived hydrogel prevents negative LV remodeling post-myocardial infarction. Biomaterials 2015, 45, 56–63. [Google Scholar] [CrossRef] [Green Version]
- Savi, M.; Bocchi, L.; Fiumana, E.; Karam, J.-P.; Frati, C.; Bonafè, F.; Cavalli, S.; Morselli, P.G.; Guarnieri, C.; Caldarera, C.M.; et al. Enhanced engraftment and repairing ability of human adipose-derived stem cells, conveyed by pharmacologically active microcarriers continuously releasing HGF and IGF-1, in healing myocardial infarction in rats. J. Biomed. Mater. Res. Part A 2015, 103, 3012–3025. [Google Scholar] [CrossRef] [Green Version]
- Lai, N.C.; Tang, T.; Gao, M.H.; Saito, M.; Miyanohara, A.; Hammond, H.K. Improved Function of the Failing Rat Heart by Regulated Expression of Insulin-Like Growth Factor I via Intramuscular Gene Transfer. Hum. Gene Ther. 2012, 23, 255–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellison, G.; Torella, D.; Dellegrottaglie, S.; Pérez-Martínez, C.; de Prado, A.P.; Vicinanza, C.; Purushothaman, S.; Galuppo, V.; Iaconetti, C.; Waring, C.D.; et al. Endogenous Cardiac Stem Cell Activation by Insulin-Like Growth Factor-1/Hepatocyte Growth Factor Intracoronary Injection Fosters Survival and Regeneration of the Infarcted Pig Heart. J. Am. Coll. Cardiol. 2011, 58, 977–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boukouaci, W.; Lauden, L.; Siewiera, J.; Dam, N.; Hocine, H.R.; Khaznadar, Z.; Tamouza, R.; Borlado, L.R.; Charron, D.; Jabrane-Ferrat, N.; et al. Natural killer cell crosstalk with allogeneic human cardiac-derived stem/progenitor cells controls persistence. Cardiovasc. Res. 2014, 104, 290–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauden, L.; Boukouaci, W.; Borlado, L.R.; López, I.P.; Sepúlveda, P.; Tamouza, R.; Charron, D.; Al-Daccak, R. Allogenicity of Human Cardiac Stem/Progenitor Cells Orchestrated by Programmed Death Ligand 1. Circ. Res. 2013, 112, 451–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Mauricio, G.; Moscoso, I.; Martín-Cancho, M.-F.; Crisóstomo, V.; Prat-Vidal, C.; Báez-Díaz, C.; Sánchez-Margallo, F.M.; Bernad, A. Combined administration of mesenchymal stem cells overexpressing IGF-1 and HGF enhances neovascularization but moderately improves cardiac regeneration in a porcine model. Stem Cell Res. Ther. 2016, 7, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samper, E.; Flores, J.M.; Blasco, M.A. Restoration of telomerase activity rescues chromosomal instability and premature aging in Terc −/− mice with short telomeres. EMBO Rep. 2001, 2, 800–807. [Google Scholar] [CrossRef] [Green Version]
- Ott, H.C.; Matthiesen, T.S.; Goh, S.-K.; Black, L.; Kren, S.; Netoff, T.; A Taylor, D. Perfusion-decellularized matrix: Using nature’s platform to engineer a bioartificial heart. Nat. Med. 2008, 14, 213–221. [Google Scholar] [CrossRef]
- Sanchez, P.L.; Santos, M.E.F.; Costanza, S.; Climent, A.M.; Moscoso, I.; Gonzalez-Nicolas, M.A.; Sanz-Ruiz, R.; Rodríguez, H.; Kren, S.M.; Garrido, G.; et al. Acellular human heart matrix: A critical step toward whole heart grafts. Biomaterials 2015, 61, 279–289. [Google Scholar] [CrossRef]
- Torán, J.L.; Lopez, J.A.; Gomes-Alves, P.; Aguilar, S.; Torroja, C.; Trevisan-Herraz, M.; Moscoso, I.; Sebastião, M.J.; Serra, M.; Brito, C.; et al. Definition of a cell surface signature for human cardiac progenitor cells after comprehensive comparative transcriptomic and proteomic characterization. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef]
- Moscoso, I.; Tejados, N.; Barreiro, O.; Sepulveda, P.; Izarra, A.; Calvo, E.; Dorronsoro, A.; Salcedo, J.M.; Sadaba, R.; Diez-Juan, A.; et al. Podocalyxin-like protein 1 is a relevant marker for human c-kitposcardiac stem cells. J. Tissue Eng. Regen. Med. 2013, 10, 580–590. [Google Scholar] [CrossRef]
- Naik, U.; Naik, M.; Eckfeld, K.; Martin-DeLeon, P.; Spychala, J. Characterization and chromosomal localization of JAM-1, a platelet receptor for a stimulatory monoclonal antibody. J. Cell Sci. 2001, 114, 539–547. [Google Scholar] [CrossRef]
- Sobocka, M.B.; Sobocki, T.; Babinska, A.; Hartwig, J.H.; Li, M.; Ehrlich, Y.H.; Kornecki, E. Signaling Pathways of the F11 Receptor (F11R; a.k.a. JAM-1, JAM-A) in Human Platelets: F11R Dimerization, Phosphorylation and Complex Formation with the Integrin GPIIIa. J. Recept. Signal Transduct. 2004, 24, 85–105. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Ji, S.; Xiao, S.; Kong, Z.; Fang, H.; Zhang, Y.; Ji, K.; Zheng, Y.; Liu, H.; Xia, Z. JAM-A promotes wound healing by enhancing both homing and secretory activities of mesenchymal stem cells. Clin. Sci. 2015, 129, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Wu, M.; Guo, X.; Liu, H. RNA interference mediated JAM-A gene silencing promotes human epidermal stem cell proliferation. Hum. Cell 2014, 28, 73–80. [Google Scholar] [CrossRef]
- Kobayashi, I.; Kobayashi-Sun, J.; Kim, A.D.; Pouget, C.; Fujita, N.; Suda, T.; Traver, D. Jam1a–Jam2a interactions regulate haematopoietic stem cell fate through Notch signalling. Nature 2014, 512, 319–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, S.; Bieda, M.C. Differences among brain tumor stem cell types and fetal neural stem cells in focal regions of histone modifications and DNA methylation, broad regions of modifications, and bivalent promoters. BMC Genom. 2014, 15, 724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izarra, A.; Moscoso, I.; Levent, E.; Cañón, S.; Cerrada, I.; Diez-Juan, A.; Blanca, V.; Núñez-Gil, I.-J.; Valiente, I.; Ruíz-Sauri, A.; et al. miR-133a Enhances the Protective Capacity of Cardiac Progenitors Cells after Myocardial Infarction. Stem Cell Rep. 2014, 3, 1029–1042. [Google Scholar] [CrossRef]
- Sakamoto, D.; Takagi, T.; Fujita, M.; Omura, S.; Yoshida, Y.; Iida, T.; Yoshimura, S. Basic Gene Expression Characteristics of Glioma Stem Cells and Human Glioblastoma. Anticancer Res. 2019, 39, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Xing, Y.; Tian, T.; Guo, Y.; Qian, J. Overexpression of LRRC59 Is Associated with Poor Prognosis and Promotes Cell Proliferation and Invasion in Lung Adenocarcinoma. OncoTargets Ther. 2020, 13, 6453–6463. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Krause, D.S.; Deans, R.J.; Keating, A.; Prockop, D.J.; Horwitz, E.M. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Adamiak, M.; Bujko, K.; Brzezniakiewicz-Janus, K.; Kucia, M.; Ratajczak, J.; Ratajczak, M.Z. The Inhibition of CD39 and CD73 Cell Surface Ectonucleotidases by Small Molecular Inhibitors Enhances the Mobilization of Bone Marrow Residing Stem Cells by Decreasing the Extracellular Level of Adenosine. Stem Cell Rev. Rep. 2019, 15, 892–899. [Google Scholar] [CrossRef] [Green Version]
- Ghalamfarsa, G.; Kazemi, M.H.; Mohseni, S.R.; Masjedi, A.; Hojjat-Farsangi, M.; Azizi, G.; Yousefi, M.; Jadidi-Niaragh, F. CD73 as a potential opportunity for cancer immunotherapy. Expert Opin. Ther. Targets 2018, 23, 127–142. [Google Scholar] [CrossRef]
- Piecewicz, S.M.; Pandey, A.; Roy, B.; Xiang, S.H.; Zetter, B.R.; Sengupta, S. Insulin-Like Growth Factors Promote Vasculogenesis in Embryonic Stem Cells. PLoS ONE 2012, 7, e32191. [Google Scholar] [CrossRef] [Green Version]
- Gong, H.; Wang, X.; Wang, L.; Liu, Y.; Wang, J.; Lv, Q.; Pang, H.; Zhang, Q.; Wang, Z. Inhibition of IGF-1 receptor kinase blocks the differentiation into cardiomyocyte-like cells of BMSCs induced by IGF-1. Mol. Med. Rep. 2017, 16, 787–793. [Google Scholar] [CrossRef] [Green Version]
- Sauer, H.; Rahimi, G.; Hescheler, J.; Wartenberg, M. Role of reactive oxygen species and phosphatidylinositol 3-kinase in cardiomyocyte differentiation of embryonic stem cells. FEBS Lett. 2000, 476, 218–223. [Google Scholar] [CrossRef]
- Zhang, G.-W.; Gu, T.-X.; Guan, X.-Y.; Sun, X.-J.; Qi, X.; Li, X.-Y.; Wang, X.-B.; Lv, F.; Yu, L.; Jiang, D.-Q.; et al. HGF and IGF-1 promote protective effects of allogeneic BMSC transplantation in rabbit model of acute myocardial infarction. Cell Prolif. 2015, 48, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Zwetsloot, P.P.; Végh, A.M.D.; Jansen of Lorkeers, S.J.J.O.; van Hout, G.P.J.; Currie, G.L.; Sena, E.S.; Gremmels, H.; Buikema, J.W.; Goumans, M.-J.; Macleod, M.R.; et al. Cardiac Stem Cell Treatment in Myocardial Infarction: A systematic review and me-ta-analysis of preclinical studies. Circ. Res. 2016, 118, 1223–1232. [Google Scholar] [CrossRef] [PubMed]






| Protein | Description | Others | Type | Log FC pCPC/hCPC | RU pCPC |
|---|---|---|---|---|---|
| Membrane | |||||
| F11R | Junctional adhesion molecule A | CD321/JAM1 | Int M | 2.45 | 16.3 |
| IL1R1 | Interleukin 1 receptor, type I | IL-1R-alpha | TK-R | 1.07 | 182 |
| IGF2R | IGF Cation-independent mannose-6-phosphate receptor | CD222/M6P-R | Mb-R | 0.31 | 16.1 |
| DPP4 | Dipeptidyl peptidase 4 | CD26 | Int M Gly | −0.38 | 190.2 |
| CACNG7 | Calcium Voltage-Gated Channel Auxiliary Subunit Gamma 7 | TARP Gamma-7 | TM | −0.98 | 42.5 |
| ECE1 | Endothelin-Converting Enzyme 1 | ECE | Enz | −1.6 | 157 |
| GPR4 | G Protein-Coupled Receptor 4 | G-PCR 19 | TM | −7.74 | 0.21 |
| Secretome | |||||
| IL1B | Interleukin 1 Beta | IL-1 Beta | Cyt | 9.1 | 100.4 |
| IGF1 | Insulin-Like Growth Factor 1 | - | GF | 8.77 | 55 |
| IL1A | Interleukin 1 Alpha | Hematopoietin-1 | Cyt | −1.81 | 122 |
| TGF b1 | Transforming Growth Factor Beta 1 | TGF-Beta-1 | GF | −2.76 | 1565 |
| Cytoplasm | |||||
| PAPSS2 | 3′-Phosphoadenosine 5′-Phosphosulfate Synthase 2 | Adeno 5-Phosphosulfate Kinase | kin | 2.77 | 1889 |
| P4HA1 | Prolyl 4-Hydroxylase Subunit Alpha 1 | P4HA | Hydrox | 1.22 | 1423 |
| PHD1 | Prolyl Hydroxylase Domain-Containing Protein 1 | EGLN2 | Hydrox | −0.42 | 81.2 |
| Nuclear | |||||
| IGF2BP2 | Insulin-Like Growth Factor 2 MRNA Binding Protein 2 | IMP2 | RNA-BP | 0.05 | 175.8 |
| IGF2BP3 | IGF2 MRNA-Binding Protein 3 | IMP3 | RNA-BP | 0.42 | 71.4 |
| GATA4 | GATA Binding Protein 4 | - | TF | −0.3 | 44 |
| WT1 | Wilms Tumor 1 | WR33 | TF | −0.91 | 6.29 |
| Groups | Group 1 (Treatment, n = 9) | Group 2 (Control, n = 7) | ||
|---|---|---|---|---|
| 1 Week (Preinjection) | 10 Weeks | 1 Week (Preinjection) | 10 Weeks | |
| LVEF (%) | 28 ± 5 | 26 ± 12 | 23 ± 7 | 19 ± 5 |
| LVEDVi (mL/m2) | 93 ± 15 | 106 ± 41 | 113 ± 20 | 122 ± 25 |
| LVESVi (mL/m2) | 67 ± 15 | 82 ± 42 | 86 ± 18 | 99 ± 26 |
| Infarct Size (%) | 20 ± 4 | 14 ± 3 | 24 ± 4 | 15 ± 4 |
| Δ LVEF (%) | n/a | −2.2 ± 10 | n/a | −3.7 ± 6.9 |
| Δ LVEDVi (mL/m2) | n/a | 12.8 ± 31 | n/a | 9 ± 22.8 |
| Δ LVESVi (mL/m2) | n/a | 15.2 ± 30 | n/a | 12.2 ± 21.2 |
| Δ Infarct Size (%) | n/a | −6.7 ± 3.3 | n/a | −9.7 ± 5.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prat-Vidal, C.; Crisóstomo, V.; Moscoso, I.; Báez-Díaz, C.; Blanco-Blázquez, V.; Gómez-Mauricio, G.; Albericio, G.; Aguilar, S.; Fernández-Santos, M.-E.; Fernández-Avilés, F.; et al. Intracoronary Delivery of Porcine Cardiac Progenitor Cells Overexpressing IGF-1 and HGF in a Pig Model of Sub-Acute Myocardial Infarction. Cells 2021, 10, 2571. https://doi.org/10.3390/cells10102571
Prat-Vidal C, Crisóstomo V, Moscoso I, Báez-Díaz C, Blanco-Blázquez V, Gómez-Mauricio G, Albericio G, Aguilar S, Fernández-Santos M-E, Fernández-Avilés F, et al. Intracoronary Delivery of Porcine Cardiac Progenitor Cells Overexpressing IGF-1 and HGF in a Pig Model of Sub-Acute Myocardial Infarction. Cells. 2021; 10(10):2571. https://doi.org/10.3390/cells10102571
Chicago/Turabian StylePrat-Vidal, Cristina, Verónica Crisóstomo, Isabel Moscoso, Claudia Báez-Díaz, Virginia Blanco-Blázquez, Guadalupe Gómez-Mauricio, Guillermo Albericio, Susana Aguilar, María-Eugenia Fernández-Santos, Francisco Fernández-Avilés, and et al. 2021. "Intracoronary Delivery of Porcine Cardiac Progenitor Cells Overexpressing IGF-1 and HGF in a Pig Model of Sub-Acute Myocardial Infarction" Cells 10, no. 10: 2571. https://doi.org/10.3390/cells10102571